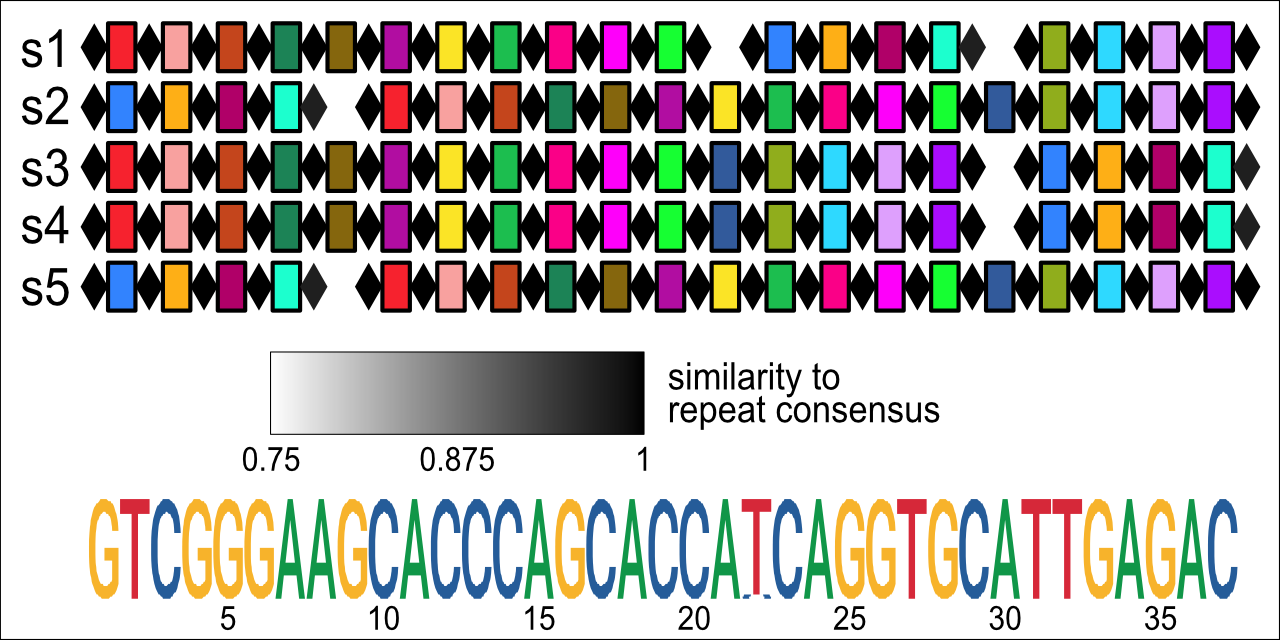
CRISPR arrays with a shared consensus repeat across five oral microbiome samples, vizualized with CRISPRviewR.
CRISPR-Cas systems are ubiquitous in environmental microbes and employ mechanistically diverse enzymes. At the heart of these systems are the arrays of spacers and repeats that serve as the basis for acquired immunity in bacteria and archaea. When analyzing complex microbial communities, a common approach to associate CRISPR-targeted mobile elements with their host species is to align spacers to viral databases or mobilome assemblies, exploiting the molecular fingerprints of past infections. To that end, many studies rely on software to identify and extract CRISPR arrays from metagenomic assemblies, generally by first identifying stretches of repeats with high sequence conservation. One popular choice is CRISPR Recognition Tool (CRT), which, despite being published in 2007, is still cited regularly, with 67 citing articles published so far in 2022. The continued popularity of CRT can likely be attributed to minCED, a fast and simple CRISPR mining tool that uses CRT under the hood.
Recently, when running minCED for a set of assemblies, I noticed something strange about one of the arrays.
View Code: minCED parameters
MIN=/path/to/minced
${MIN}/minced \
-gffFull \
-minNR 3 \
-minRL 20 \ # default: 23
-maxRL 50 \ # default: 47
-minSL 22 \ # default: 26
-maxSL 55 \ # default: 50
contigs.fasta \
example.txt \
example.gffCRISPR 1 Range: 125549 - 126048
POSITION REPEAT SPACER
-------- -------------------------------- ----------------------------------------------
125549 GTTGTCATTAGCTTCCAGATTCCGTACCTTCA CACTTGCTAATACAGCTGTGGTTGAGCCAAACAATGAGATGGTAAT [ 32, 46 ]
125627 GTTGTGATTAGCTTTCAGATTCCGTACCTTCA TACTTGCTAATACAGCGCACGCGAGACCTTCACGCGACTAGGACGG [ 32, 46 ]
125705 GTTGTGATTAGCTTTCAGATTCCGTACCTTCA TACTTGCTAATACAGCCACGAGCCTCATCACGCGAACTCTCATCAC [ 32, 46 ]
125783 GTTGTGATTAGCTTTCAGATTCCGCACCTTCA TACTTGCTAATCCAGCCGAATTATTGCAACGCTTATCCTCGCCTCG [ 32, 46 ]
125861 GTTGTGATTAGCTTTCGAATTCCGTACCTTCA CACTTGCTAACACAGCATAAAAACGACGACGACACGACCGACAGGT [ 32, 46 ]
125939 GTTGTGATTAGCTTTCAGATTCCGTACCTTCA CACTTGCTAATACAGCTCGGAGGAGTGAAGAATAGCCAGCACCTCG [ 32, 46 ]
126017 GTTGTGATTAGCTTTCAGATTCCGTACCTCCA
-------- -------------------------------- ----------------------------------------------
Repeats: 7 Average Length: 32 Average Length: 46Specifically, the first 16 nt are more similar between spacers than expected by chance. Obviously, some portion of the repeats are being misassigned to the spacers. This discrepancy does not appear to be the result of bad parameterization, as the expected lengths for the repeats and spacers (48 and 30 nt, respectively) fall within the ranges given in the parameters. So, I opened an issue on GitHub, and Connor Skennerton, the creator of minCED, soon replied with an explanation:
It is a limitation in the core CRT algorithm where it expects the repeats to be identical. In repeats where there is a slight wiggle the algorithm messes things up, as you have observed. There is a magic number in the core repeat extension algorithm where 75% of the nucleotides need to be the same to get to extend to the next base …
As minCED depends on CRT, other tools likewise depend on minCED, including CRISPRdisco and CRISPR Visualizer, and thus the inherent limitation of CRT is propagated. Instead of learning Java and fixing CRT at the source, I opted to implement a post-hoc correction in R as a function in my CRISPRviewR package.
If fix_repeats = TRUE is specified, read_minced() will attempt to reassign portions of spacers to repeats by calculating rolling average conservation scores across spacer consensus sequences representing both possible array orientations. Reading from the left end of the conservation score vector, a new spacer-repeat boundary is drawn at the position where the conservation score falls below the threshold value given by cutoff. When cutoff = 0.7 and window = 2 (defaults), this method reliably reassigns more-similar-than-expected spacer substrings to their neighboring repeats. In a test set of minCED data from 5 oral microbiome assemblies, more than 10% of annotated CRISPR arrays (109 of 964) were found to have truncated repeats.
View Code: read minCED data, original
library(CRISPRviewR) # v0.2.0
baseurl <- "https://raw.githubusercontent.com/acvill/CRISPRviewR/master/example_data_minced/"
cdat <- merge_minced(
s1 = read_minced(txt = url(paste0(baseurl, "s1.txt")),
gff = url(paste0(baseurl, "s1.gff"))),
s2 = read_minced(txt = url(paste0(baseurl, "s2.txt")),
gff = url(paste0(baseurl, "s2.gff"))),
s3 = read_minced(txt = url(paste0(baseurl, "s3.txt")),
gff = url(paste0(baseurl, "s3.gff"))),
s4 = read_minced(txt = url(paste0(baseurl, "s4.txt")),
gff = url(paste0(baseurl, "s4.gff"))),
s5 = read_minced(txt = url(paste0(baseurl, "s5.txt")),
gff = url(paste0(baseurl, "s5.gff")))
)View Code: read minCED data, fixed
cdat_fixed <- merge_minced(
s1 = read_minced(txt = url(paste0(baseurl, "s1.txt")),
gff = url(paste0(baseurl, "s1.gff")),
fix_repeats = TRUE),
s2 = read_minced(txt = url(paste0(baseurl, "s2.txt")),
gff = url(paste0(baseurl, "s2.gff")),
fix_repeats = TRUE),
s3 = read_minced(txt = url(paste0(baseurl, "s3.txt")),
gff = url(paste0(baseurl, "s3.gff")),
fix_repeats = TRUE),
s4 = read_minced(txt = url(paste0(baseurl, "s4.txt")),
gff = url(paste0(baseurl, "s4.gff")),
fix_repeats = TRUE),
s5 = read_minced(txt = url(paste0(baseurl, "s5.txt")),
gff = url(paste0(baseurl, "s5.gff")),
fix_repeats = TRUE)
)An analysis of the sequence content of the 109 fixed arrays shows that, on average, 8 nt from spacers are reassigned to repeats. The most important windfall of correctly annotating the spacer-repeat boundary is more accurate host-phage pair assignment. Though not tested here, alignments of spacers to phage databases often rely on BLAST searches optimized for short sequences, and such alignment schemes are likely affected by excess non-spacer sequence.
View Code: summarize fixed sequences
library(tidyverse) # v1.3.2
compare_rep <- function(dat) {
dat |> group_by(id, array) |> slice(2) |> select(id, array, rep)
}
comp <- left_join(compare_rep(cdat),
compare_rep(cdat_fixed),
by = c("id", "array")) |>
filter(rep.x != rep.y) |>
mutate(diff = nchar(rep.y) - nchar(rep.x))
ggplot(data = comp) +
geom_histogram(mapping = aes(x = diff),
binwidth = 1,
color = "blue",
fill = "cornsilk") +
geom_vline(mapping = aes(xintercept = mean(diff)),
color = "red",
linetype = "dashed",
size = 1) +
scale_x_continuous(breaks = seq(1, 25, by = 2)) +
xlab("nt added to fixed repeats") +
ylab("count of CRISPR arrays") +
theme(axis.title.x = element_text(size = 15),
axis.title.y = element_text(size = 15),
axis.text.x = element_text(size = 13, angle = 0,
vjust = 0.5, color = "black"),
axis.text.y = element_text(size = 13, color = "black"),
panel.border = element_rect(color = "black", linewidth = 1.5,
fill = NA, linetype = "solid"),
panel.background = element_rect(fill = "lightblue"),
axis.line.y = element_blank(),
axis.line.x = element_blank(),
panel.grid = element_blank(),
axis.line = element_line(color = "black"))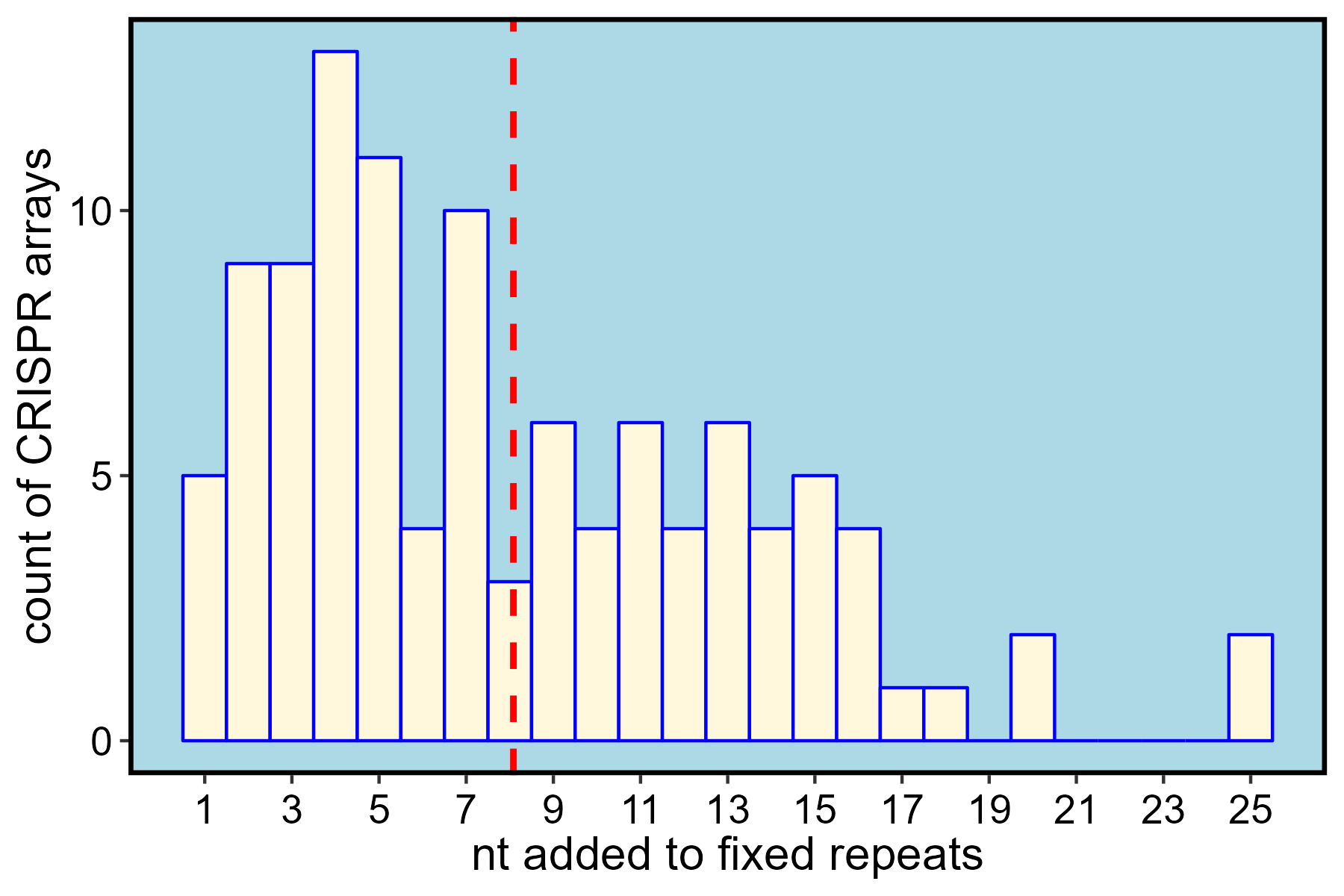
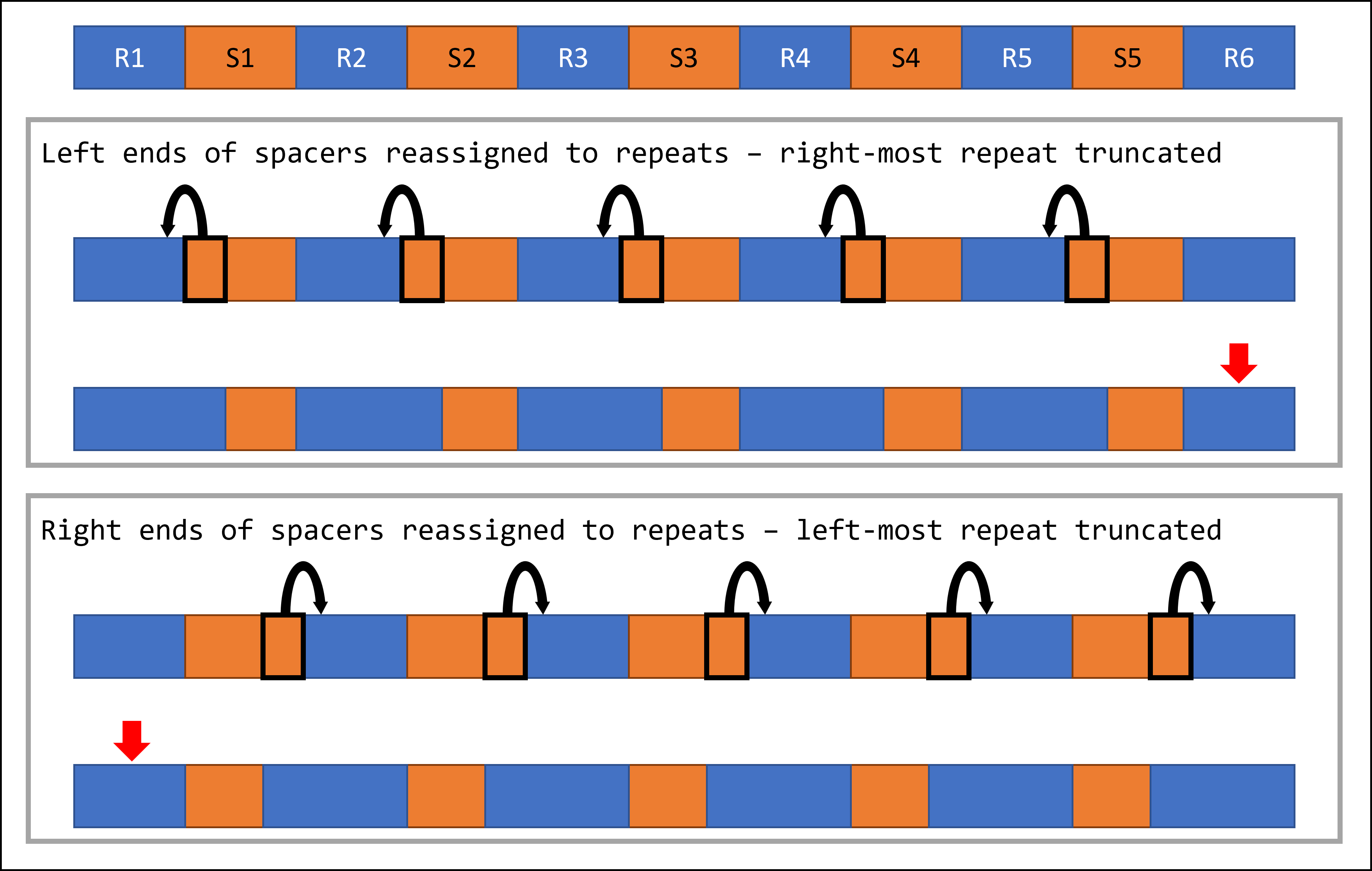
One important caveat of this approach is that it is agnostic to the sequence context of the original assembly, since it relies solely on the minCED output as input. This means that one repeat at one end of each fixed array will retain its original truncated sequence, as detailed in the schematic below.