I upload standalone functions and short scripts to my GitHub Gists page.
Expand details…
minCED (Mining CRISPRs in Environmental Datasets) is a program used by prokka and other pipelines for the annotation of CRISPRs in metagenomic assemblies. However, manually parsing the output of minCED for interesting CRISPRs is tedious, and comparing CRISPRs across samples is difficult because of differences in array assembly and orientation. CRISPRviewR attempts to address this issue by associating CRISPR arrays by shared consensus repeat sequences, permitting clustering and visualization of spacers between metagenomic assemblies. This package also includes a function to fix truncated repeats. Check out the CRISPRviewR vignette for use cases and example plots.

Expand details…
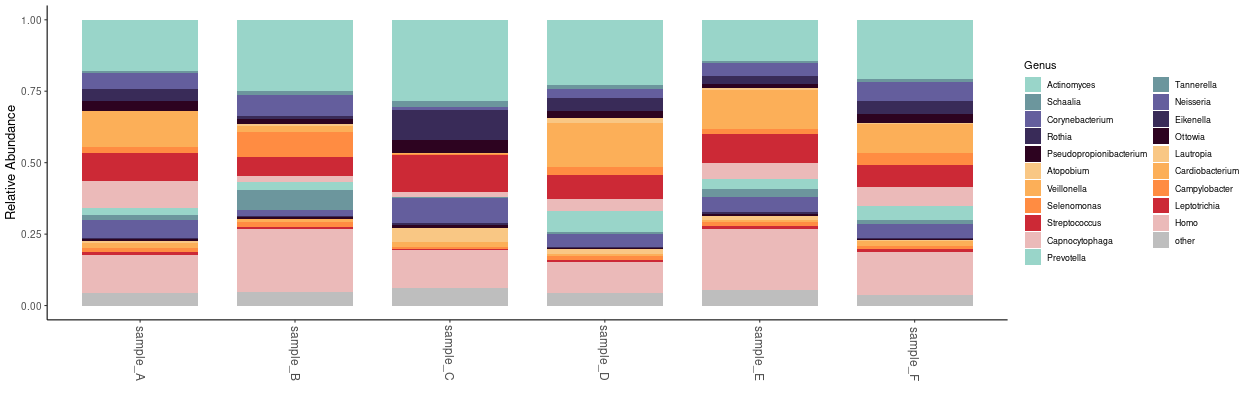
I frequently use Derrick Wood’s Kraken2 software for accurate and lightning-fast classification of short reads. Bracken is complimentary software that uses Kraken reports to produce relative abundance estimates at any taxonomic level. Often, a first step in metagenomic analysis is examining the distribution of different organisms across samples. To that end, I’ve created a simple shiny app for quick, customizable plotting of taxonomic structures encoded in multi-sample Bracken reports.
Expand details…
nodeSeqs.sh takes in a Graphical Fragment Assembly file and outputs sequences proximal to high-degree nodes. In the language of GFA, this script looks for segments that have a large number of links, and then filters those segments by k-mer coverage to preclude hits that are poorly assembled due to low coverage. The resulting .fasta can then be fed into your favorite sequence alignment software.
For an example application of this script, please see my blog post: Lost at the crossroads: genes at the nodes of short-read assembly graphs.

Expand details…
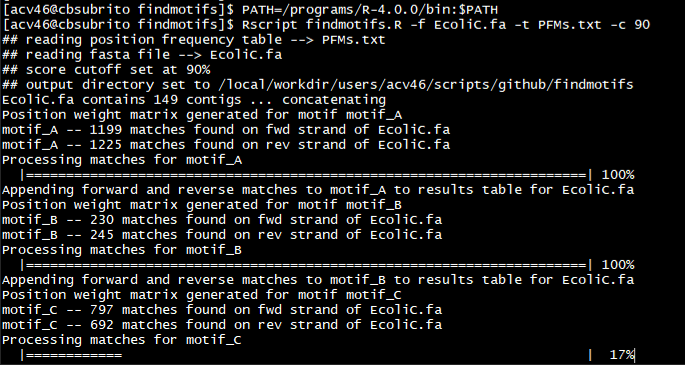
To understand the genetic context and biological relevance of DNA motifs, it’s often valuable to analyze their distribution and sequence conservation. Many programs exist for both the identification of novel motifs and the scoring of known motifs, though few of these tools work out-of-the-box for highly fragmented bacterial genomes. To address this, I’ve written findMotifs.R – an easy-to-implement R script to find and score short sequences using position-frequency matrices (PFMs). Given a set of contigs and a list of PFMs, this script returns an easy-to-parse table containing the sequence, score, and strand-wise position of each match above a user-defined threshold. findMotifs.R requires R version 4.0 and three packages: Biostrings, optparse, and stringr.